Paper: Lu, M. Y., Chen, T. Y., Williamson, D. F., Zhao, M., Shady, M., Lipkova, J., & Mahmood, F. (2021). AI-based pathology predicts origins for cancers of unknown primary. Nature, 594(7861), 106-110.
Available Materials
- Data
- TCGA diagnostic WSI data and corresponding labels: NIH genomic data commons
- CPTAC histology data and corresponding labels:TCIA CPTAC Pathology Portal
- Email all requests for academic use of raw and processed data to the corresponding author (and also include M.Y.L. (mlu16@bwh.harvard.edu)).
- Code: mahmoodlab/TOAD
Abstract
- Cancer of unknown primary(CUP) origin: an enigmatic group of diagnoses in which the primary anatomical site of tumour origin cannot be determined.
- Usually solved by
- genomics: lacks clinical penetration in low-resource settings
- transcriptomics
- Propose: Tumor Origin Assessment via Deep Learning(TOAD)
- provide a differential diagnosis for the origin of the primary tumor
- using routinely acquired histology slides
- Method
- Training Data: Whole-slide images(WSI) with known primary origins
- Model: simultaneously identifies the tumour as primary or metastatic and predicts its site of origin.
- Result
- held-out test set: top-1 accuracy of 0.83 and a top-3 accuracy of 0.96
- external test set: top-1 and top-3 accuracies of 0.80 and 0.93
- further curated dataset of 317 cases of CUP for which a differential diagnosis was assigned: in concordance for 61% of cases and a top-3 agreement of 82%.
- Usually solved by
TOAD workflow
Data
- 32537 gigapixel WSIs(22833 WSIs were used for training, 6499 WSIs were held out for test)
- spanning 18 groups of common primary cancer origins
- 25,419 primary and 7,118 metastatic WSIs from 29,107 patient; 52.8% women, 47.2% men)
- All WSIs were processed and analysed at the 20× equivalent magnification.
- The dataset is randomly partitioned and is stratified by class, into a training set (70% of cases), a validation set (10% of cases) and a test set (20% of cases)
- Data sources
- Publicly available data: In total, we gathered 10,406 WSIs from 8,794 patients across 25 TCGA studies and 2,969 WSIs from 1,151 patients across 7 CPTAC studies
- In-house data: 19,162 WSIs (12,215 primary, 6,947 metastatic) from internal patients at the Brigham and Women’s Hospital
- External test set: 682 samples from more than 200 medical centres
- Additional CPU test dataset: 317 cases of CUP that were assigned a primary differential based on ancillary tests, radiology, patient history, clinical correlation or at autopsy
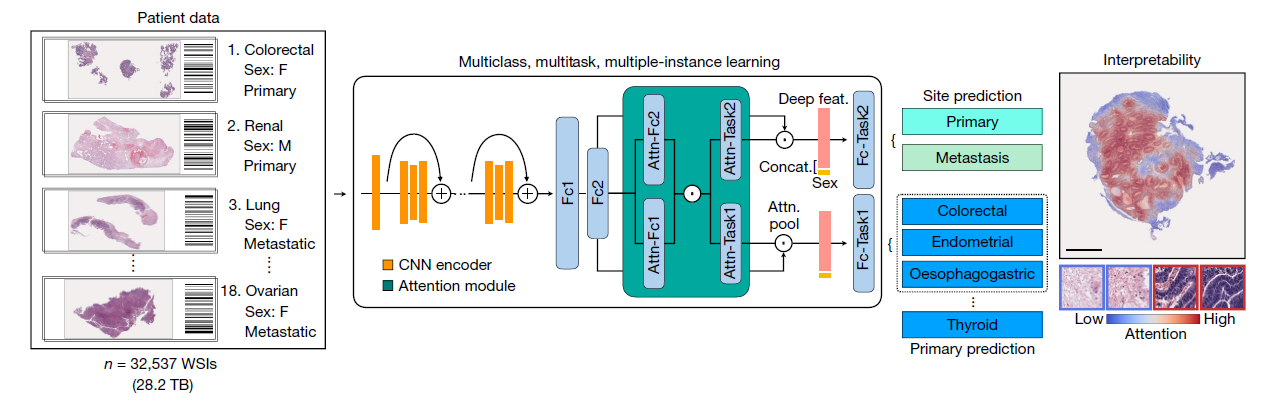
TOAD model architecture
-
Purpose: to simultaneously predict the origin of the tumour in each WSI and whether it was the primary tumour or a metastasis.
-
Technique: a form of weakly supervised machine learning known as multiple instance learning
- considering each WSI as a collection (known as a bag) of smaller image regions (known as instances)
-
Workflow
-
WSI processing: tissue segmentation and patching
-
Dimensionality reduction: patch –> fixed pretrained ResNet50 –> 1024-dimensional feature vector
-
Multitask attention pooling
-
Task1: Primary Prediction (18 groups), Task2: Site Prediction(Primary vs Metastatic)
- FC1: $W_1\in\mathbb{R}^{512\times1024}, b_1\in\mathbb{R}^{512}$
- FC2:$W_2\in\mathbb{R}^{512\times512}, b_2\in\mathbb{R}^{512}$
- $h_k=ReLU(W_2(ReLU(W_1z_k+b_1))+b_2)\in\mathbb{R}^{512}$
- $h_{slide,t}\in\mathbb{R}^{512}=\sum{a_{k,t}h_k}$, $t\in{1,2}$indicates tasks, $a_{k,t}$indicates attention
- $a_{k,t}=\frac{exp{W_{a,t}(tanh(V_ah_k))\odot sigm(U_ah_k)}}{\sum_{j=1}^N{exp{W_{a,t}(tanh(V_ah_j))\odot sigm(U_ah_j)}}}$
-
- Late-stage fusion and classification
- incorporate the biological sex of each patient into the prediction
- $p_t=softmax(W_{cls,t}concat([h_{slide,t},s])+b_{cls,t})$
- Multi-task Loss
- $\mathcal{L}{total}=c_1\mathcal{L}{cls,1}+c_2\mathcal{L}_{cls,2}$
- $c_1=0.75, c_2=0.25$ give better performance
-
Evaluation of model performance
-
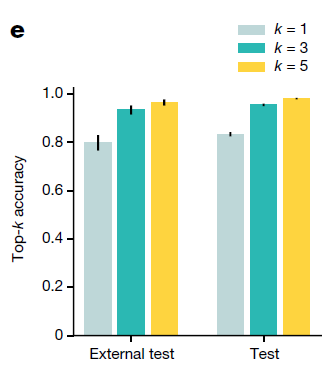
Top-k Performance
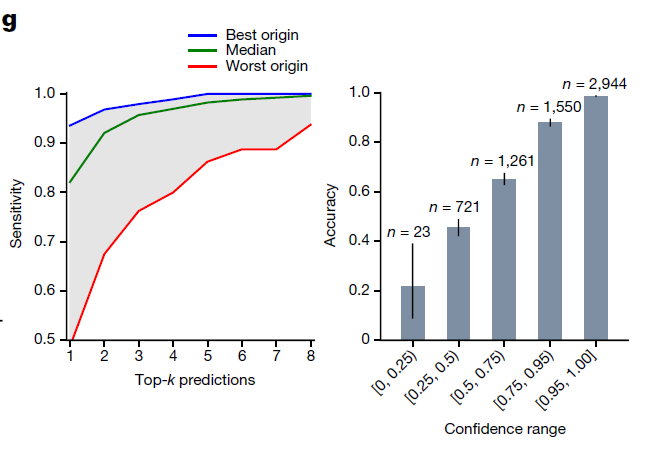
The high top-k accuracy suggests that we can potentially use the top predictions of TOAD for a given slide to narrow down the origin of the tumor to a handful of possible origins
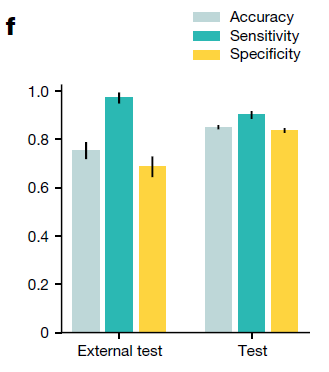
TOAD was able to predict whether the tumour specimen is a metastasis with an accuracy of 85.0% and AUC of 0.942
-
Primary versus metastasis performance
-
Generalization to external test cohort
Without tuning or domain adaptation, our trained model produced an accuracy of 79.9% and a top-3 accuracy of 93.4%
Similarly, on the second task of distinguishing between a metastasis and a primary tumour, the model had an AUC of 0.919
-
Confusion Matrix on held-out test set
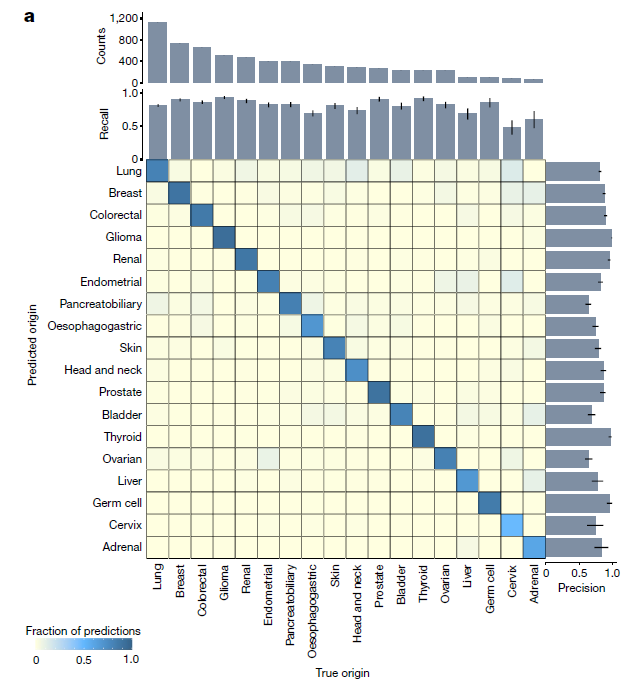
-
fraction of samples that was correctly classified
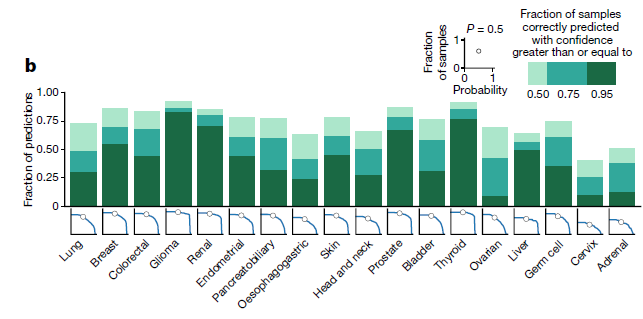
-
Micro-averaged one-versus-rest ROC(Left) and primary versus metastasis ROC(Right)
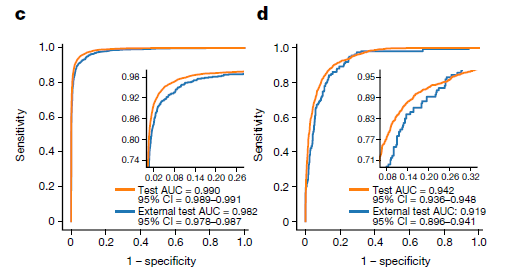
-
Further analysis of the model predictions on the test set (n = 6,499)